3D Modeling in Biology
HISTORY
1951: Linus Pauling & Robert Corey
Pauling and Corey of the California Institute of Technology took part in some of the first research on amino acids and how they link together to form chains and complex bundles. Together, they used their experience in math, biology, and model making, to understand the 3-dimensional formations of chemicals bonds within protein complexes. Two basic yet fundamental 3D structures discovered by Pauling and Corey are the alpha helix and beta sheet that are present in folded proteins.


1953: Discovery of Double Helix
Although attributed to Watson and Crick, many scientists contributed to the discovery of the sturctural model of the DNA molecule. Before the discovery, researches in the field realized that uncovering what DNA looked like was the key to moving forward with the understanding of cell biology. Based on crystalography research conducted by Maurice Wilkins and Rosalind Franklin, as well as a chemical structure discovered by Alexander Todd, Watson and Crick were able to create an accurate model of DNA, which was an important step for the advancement of biological research.
1973: Chernoff Faces
Herman Chernoff, a professor of Applied Mathematics at MIT and Statistics at Harvard University, published a monograph called “The Use of Faces to Represent Points in K-Dimensional Space Graphically." He used drawings of human faces to portray statistical data. By converting data into recognizable images, Chernoff demonstrated that graphical models are immensly helpful in analysing data.

1978: Nelson Max
Nelson Max was a teacher of math and graphical modeling at UC Davis during the 1970s and 80s. Max was instrumental in bringing computer animation to both entertainment and science. In 1978, he completed a now famous computer generated 3D vizualization of a DNA Molecule.
1987: Bill Lorensen & Harvey Cline
Bill Lorensen and Harvey Cline, while working at General Electric, researched ways to efficiently visualize data from MRI and CT scans. They developed an algorithm called "Marching Cubes," which was the foundation of computer generated imagery. Applications of this algorithm used extensively in the biomedical field.


1987: apE Software
Animation Production Environment, apE, was the first publicly available software for 3D modeling. This software was developed by researches at Ohio State University as part of the Ohio Supercomputer Graphics Project (OSGP). Its development was backed and funded in part by Apple Computer. apE was made free to the public, open source, and was the precursor for several commercial scientific visualization products.
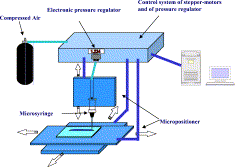
2003: 3D Tissue Scafold
In 2003, researchers from Italy published a paper on their work in 3D bioprinting. These researches were able to extrude tissue cells from a nozzle and form layers of cells in a scaffolded arrangement. This was the proof-of-concept for tissue engineering and layed the groundwork for the industry of bioprinting.